Optimising a Disease Spread Model: Presentation to the Big Data Institute
Following from our development of a performance-optimised transmission model for onchocerciasis, we were invited to visit the Big Data Institute at the University of Oxford to present our optimisations and the results.
After a review of the original model, which is written in R, we moved forward on an initial stage of translation to Python. This involved a significant refactor, in which we built a higher level representation of the model state and running steps, and which enabled us to better analyse the blocking run-time steps.

This clearer structure enabled us to make some initial optimisations, and then profile the code:
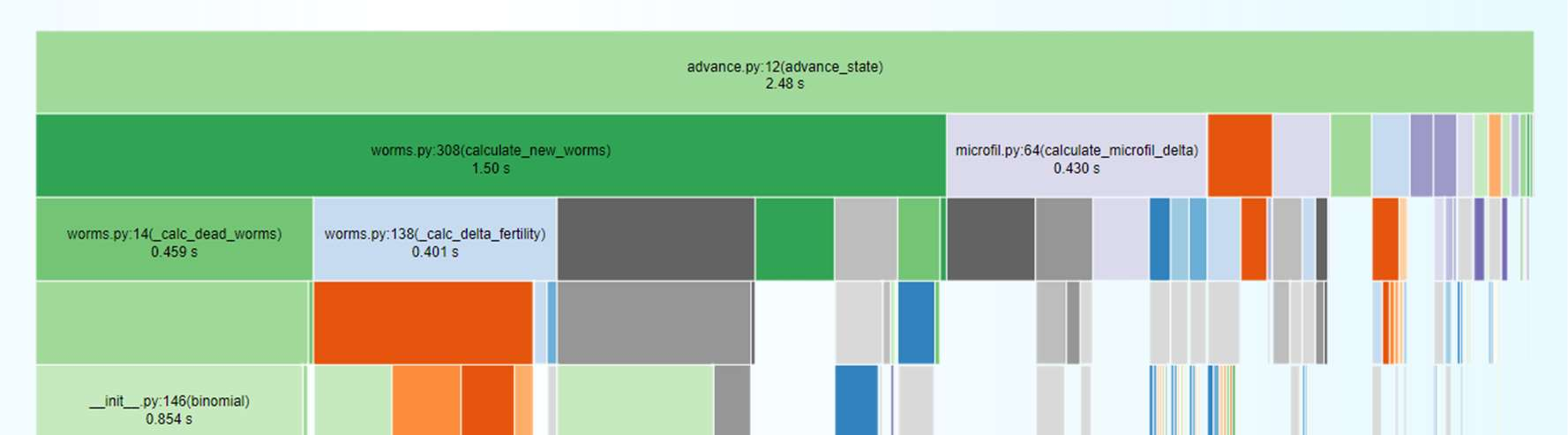
After identifying the blocking steps, we made the following optimisations:
Firstly, we removed any tight Python loop iterations, replacing them with their vectorised NumPy equivalents. This resulted in a significant speed-up!
After this optimisation, the net slowest function was the sampling of binomial distributions, which was already being implemented through NumPy. We therefore wrote a custom C++ library for fast array-optimised binomial sampling: fast-binomial. By generating the values in bulk and referencing them later in a look-up table, we achieved a further order of magnitude speed-up.
To ensure that our implementation had the same behaviour as the original R model, despite both being stochastic models, we developed pytest-trust-random, a testing suite for comparing the stochastic results between two different models. This has been adopted by the NTD modelling consortium as a core tool for model development.
Further information about this project can be found in the slides, which are available here! The final source code for the project is available here.